
생화학분자생물학회입니다.
단일 세포 다중 오믹스 분석 (Single cell multi-omics analysis)
작성자
관리자작성일자
2019-12-13조회수
4555
단일 세포 다중 오믹스 분석
(Single cell multi-omics analysis)



배서경, 김경대, 박지환
광주과학기술원 생명과학부
qotjrud3@gist.ac.kr
ruddae43@gist.ac.kr
jihwan.park@gist.ac.kr
서론
인체는 최소 200개 이상의 서로 다른 유형의 세포로 구성되어 있다고 알려져 있으며, 세포마다 독특한 특성을 가지고 있다. 서로 다른 특성을 가진 세포를 세포 유형 특이적으로 분석할 수 있는 기술인 단일세포 분석 기술은 최근 급속히 발전하고 있는 분야이다. 단일세포 분석은 수백~수천 개의 온전한 세포들을 분리하여 각각의 세포에 발현된 유전물질을 분리, 시퀀싱하여 세포 종류를 밝히고 세포 특이적인 변화를 동정할 수 있으며, 나아가 시공간적 관계까지 재구축할 수 있는 기술이다. 앞으로 생명과학 연구에 혁명을 가져올 것이라는 기대와 함께 2018년 Science 저널에서 breakthrough로 선정되었고[1], 인간의 모든 세포 유형과 위치를 밝히고, 세포들이 협력하여 조직과 기관을 형성하는 기작을 연구하고자 하는 국제적인 협력 프로젝트인 Human Cell Atlas Project가 시작되기도 하였다.
모든 세포는 하나의 zygote에서 파생되어 같은 유전자를 가지지만 모든 세포에서 같은 유전자가 발현되지 않고 다양한 세포 유형을 생성한다. 조직은 다양한 세포 집단들로 복잡하게 구성되어 있으며 동일한 세포 유형에서도 intrinsic, extrinsic variation에 의해 유전자 발현, 세포의 기능과 형태가 다를 수 있다. 이러한 세포의 특이성을 확인하고 동정하는 과정은 생물학적 과정에 대한 이해와 세포에 대한 근본적인 접근에 필수적이다. 그러나 기존의 bulk RNA-seq으로는 전체 세포 집합들의 평균적인 정보만을 관찰하기 때문에 여러 종류의 세포 유형을 구분하여 분석하는데 어려움이 있었다[2]. Single cell RNA-sequencing (scRNA-seq)은 기존의 RNA-seq과 달리 세포 유형의 구성 변화와 세포 종류 특이적인 변화를 구분하여 확인할 수 있으며, 조직 수준을 넘어 세포 유형 특이적인 정보를 얻을 수 있다. 또한, scRNA-seq은 세포 분화 및 질병 발생 과정에 대한 cell trajectory 분석을 할 수 있어, 연속적인 세포 변화과정에서의 핵심 조절 유전자를 밝혀낼 수 있다. 이와 같이 기존의 기술의 한계를 극복하고 단일 세포 수준에서 genotypic, phenotypic heterogeneity 정보를 얻을 수 있는 scRNA-seq을 이용한 연구의 중요성과 보편성은 점점 증가하고 있다.
Single cell sequencing을 이용한 연구는 전사체(transcriptome)를 분석하는 것을 넘어서 후성유전체(epigenome), 단백체(proteome), 대사체(metabolism)과 같은 다양한 분자 수준의 데이터를 분석하는 것으로 발전하고 있다. 세포의 기능은 둘 이상의 분자 유형에 의해 결정되며, 전사체만을 이용한 연구에서는 세포의 변화를 관찰하는데 일편적이고 한정된 정보를 제공하기 때문에 최종 상태에서 실제로 나타나는 형질은 전사체에서 관찰된 것과 다를 수 있다[3]. 반면, 여러 종류의 분자를 단일 세포 수준에서 측정하면 bulk sample의 heterogeneity에 의한 영향 없이 세포의 genotype과 phenotype의 연관성을 보다 정확하게 분석할 수 있다. 또한, 여러 유전자와 외부 요인들이 복합적으로 얽혀 발생하는 질병에 근본적인 접근을 하기 위해서도 단순히 하나의 요인만을 고려하기보다는 여러 요인들을 통합하여 하나의 집합체로서 접근하는 방식이 필요하다. 따라서 이 글을 통해 다양한 single cell sequencing을 이용한 오믹스 기술과 동향에 대해서 알아보고자 한다.
본론
2-1. Single cell transcriptomics
scRNA-seq은 일반적으로 단일 세포 분리, lysis, cDNA 합성, 증폭, library 준비와 sequencing 과정을 포함하며 조직이나 기관을 단일 세포로 분리하는 것으로부터 시작한다. 단일 세포를 분리하는 방법에는 Fluorescence-activated cell sorting, microfluidic platform, serial dilution 등이 있으며, 조직의 종류와 sequencing type에 따라 최적화된 방법을 선택한다[4]. 분리된 단일 세포들은 platform에 따라 다른 방식으로 RNA를 capture하며, 각 platform의 특성에 따라 처리된 RNA는 sequencing된 후 컴퓨터 처리를 통해 분석이 진행된다.
scRNA-seq은 크게 full-length scRNA-seq과 tag-based scRNA-seq으로 나뉜다. Full-length scRNA-seq은 유전자 발현, transcript isoform, alternative splicing, SNP, mutation 정보를 포함한 전사체 전체를 sequencing할 수 있는 기술로, 대표적인 방법으로는 Smart-seq2, Quartz-seq이 있다[5]. Full-length RNA-seq은 높은 coverage와 매핑 효율을 보이지만, 샘플이 독립적으로 준비되어야 하므로 처리량에 제한이 있고 실험 간 batch 효과가 상대적으로 크며, 샘플 준비 시간과 비용이 많이 든다는 단점이 있다[6]. 반면, tag-based scRNA-seq은 주로 transcript의 3’-end를 시퀀싱하여 transcript abundance를 추정하는데 사용되는 기술로, 하나의 튜브에 모든 샘플을 통합하여 sequencing library를 만든다[6]. 이를 위해 서로 다른 세포를 구별하기 위한 label과 transcript copy를 정확하게 측정하기 위한 molecular label을 primer 말단에 첨가하는데, droplet-based, microwell-based, split-pool barcoding 기술로 나뉜다. Droplet based 기술은 oil droplet과 resin, hydrogel 또는 magnetic material로 만들어진 barcoded bead를 사용하는 기술로, chamber에 물과 oil을 통해 droplet을 만들고, droplet에 하나의 세포와 하나의 bead가 쌍을 이루어 캡슐화 되어 bead의 oligo dT에 의해 각 단일 세포의 mRNA를 capture한다[6]. Droplet-based barcoding은 read길이가 제한되어 주로 barcode에 가까운 3’-end mRNA 정보를 얻는다는 한계가 있지만, 단일 세포 분석의 처리량을 향상시키고 실험 절차를 간소화하여 시간과 노력 그리고 batch 효과를 감소시켰으며, 세포 당 비용을 크게 감소시켰다. Microwell-based 기술은 105개의 microwell이 있는 plate에 세포를 로딩 하고 모세관으로 doublet을 씻어낸 후, barcoded beads를 첨가하여 mRNA를 capture하는 방법이다[7]. Microwell-based 기술은 실험 방법이 단순하며 비용이 낮다는 장점이 있지만, 분리되어 처리되어야 하며 오염의 가능성이 있다. 마지막으로 split-pool barcode는 단일세포로의 분리과정 없이 combinatorial indexing으로 단일 세포를 확인하는 방법으로, sci-RNA-seq(single-cell combinatorial indexing RNA sequencing)과 SPLiT-seq(split-pool ligation-based transcriptome sequencing)이 대표적이다. 이 기술들은 permeabilization 시킨 세포 수 십 개를 96- 또는 384-well plate에 분배하여 세포 내에서 역전사를 하면서 1차로 바코딩을 하고 다시 세포를 pooling하여 다음 well plate에 수 십 개씩 분배하여 2차 barcode를 순차적으로 추가해줌으로써 세포 별 고유한 바코드를 추가해주는 기술이다[8]. Split-pool barcode는 높은 처리량, 낮은 비용의 장점이 있다.
scRNA-seq은 샘플 준비 단계에서 세포를 분리하기 어렵거나 분리 과정에 의한 damage에 민감한 세포인 경우 분석에 어려움이 있다. Single nucleus sequencing(snRNA-seq)은 이러한 한계를 극복한 기술로, 단일 세포가 아닌 single nucleus를 분리하여 전사체 분석을 진행하는 방식이다. scRNA-seq은 fresh한 조직 샘플을 사용해야 하는 반면 snRNA-seq은 frozen sample도 사용할 수 있으며, intertwined 세포로 인해 분리가 어려운 조직의 전사체도 분석할 수 있다[9]. 또한 분리 단계에서 생기는 스트레스와 bias를 감소시킬 수 있다는 장점이 있다. 하지만 핵에 있는 RNA만을 확인할 수 있기 때문에 세포질의 RNA에 대한 정보는 얻을 수 없고, 인트론 서열이 많으며 세포 구성 성분에 bias가 생긴다는 단점이 있으므로 실험 목적에 맞게 선택할 필요가 있다.
2-2. Single cell epigenomics
세포의 유전자 서열의 변화 없이 유전자 발현에 영향을 미치는 DNA methylation, histone modification과 같은 epigenomics를 연구하기 위한 single cell sequencing 기술도 최근 등장하고 있다. Single cell DNA methylation 분석을 통합하는 접근법인 scBS-seq(single-cell bisulfite sequencing)과 scRRBS-seq(reduced representation bisulfite sequencing)이 최근 개발되어 세포 genome의 CpG site에 기반한 DNA methylation 정보를 제공하고 있다[10]. 유전자와 단백질의 상호작용을 분석하는 ChIP-seq은 ChIP(chromatin immunoprecipitation)과 sequencing을 결합한 기술로, DNA-associated protein의 결합 지역을 확인할 수 있어 histone modification과 전사 인자의 결합 지역을 확인하는데 사용된다[11]. Single cell ATAC-seq (Assay for Transposase-Accessible Chromatin)은 열린 크로마틴 지역을 매핑할 수 있는 기술이다. 이 기술은 modified Tn5 효소를 통해 열린 크로마틴 지역을 잘라냄과 동시에 adapter를 부착시키는 기술이다[12]. 아직까지는 단일 세포 수준에서는 원하는 모든 타겟에 접근할 수 없다는 점, 대량의 custom modified Tn5로 인한 높은 비용 등과 같은 문제점은 아직 풀어가야 할 숙제로 남아있지만, 세포 종류별 기능성 DNA 지역을 찾고, cell-to-cell chromatin variation의 결정 요인을 확인할 수 있을 것이다. 이외에도 단일 세포 수준에서 염색체 형태를 평가할 수 있는 기술인 single-cell Hi-C은 염색체에 대한 평균적인 정보만을 제공하는 기존 Hi-C의 한계를 극복하고, 개별 염색체의 구조, 구획화 및 염색체 간의 상호작용에 대한 정보를 주는 기술로 자리매김 하고 있다[13]. scHi-C는 genomic loci간의 potential interaction에 대해 아직 낮은 recovery rate를 보이지만, 전체 genome의 3차원 map을 만드는 것을 가능하게 하였다.
2-3. Single cell proteomics
Transcription이 항상 동일한 단백질로 나타나는 것이 아니며, 세포의 표현형을 나타내는 궁극적인 단위는 단백질이기 때문에 단백체 연구 필요성은 항상 제기되어왔다. 특히 scRNA-seq에서는 rare type의 전사체나 일정하게 전사되지 않는 전사체에서는 detection이 일정하지 않거나 없는 상태인 “dropout effect”가 자주 발생하므로, 이를 보완하기 위한 단일 세포 단백체를 분석 기술들이 개발되어왔다. Antibody를 이용한 방법으로는 기존의 flow cytometry를 이용한 CyTOF 와 fluorescence를 기반으로 한 multiplexed imaging platform인 CODEX(CO-Detection by antibody indexing)가 있다. Mass spectrometry를 이용한 ScoPe-MS(Single Cell Proteomics by Mass Spectrometry) 역시 세포 유형 및 progression을 확인하기 위한 방법으로 자리 잡고 있다[14-16]. 또한 최근 barcode oligonucleotide가 결합된 단백질 결합 분자를 이용하여 세포 표면 단백질을 labeling하고 정량화 하는 기술인 CITE-seq (Cellular Indexing of Transcriptomes and Epitopes by Sequencing) 및 REAP-seq (RNA expression and protein sequencing)과 같은 기술들이 개발되었다[17, 18]. Single cell proteome 정보는 세포의 주된 functional machinery인 단백질이 세포마다 다르게 발현되는 양상을 보다 직접적으로 확인하는데 도움을 준다.
2-4. Single cell spatial transcriptomics
세포를 보다 잘 이해하기 위한 노력은 실제에 가까운 세포 환경을 재현함으로써 지속되고 있다. 세포는 3차원 공간에 존재하며 그 공간 내에서 상호작용하고 있기 때문에 spatial transcriptome 분석을 통해 세포 종류 및 위치를 이해하기 위한 방법들이 개발되었다. 기존의 smFISH(single molecule fluorescence in situ hybridization)는 RNA의 위치 및 copy number파악을 위해 사용되었지만 많은 세포를 동시에 측정할 수 없다는 단점이 있었다. 이러한 단점을 극복하기 위해 단일 세포에서 다수의 RNA labeling하여 FISH를 하는 방법인 MERFISH(multiplexed error-robust FISH)와 FISH를 여러 번 진행하여 imaging하는 방법인 seq-FISH(sequential-FISH)가 개발되었다[19, 20]. 뿐만 아니라 10x Genomics에서 “Spatial Transcriptomics”란 기술을 서비스하고 있으며, 이외에도 DNA probe로 RNA를 labeling하는 STARmap (spatially resolved transcript amplicon readout mapping) 및 기존의 padlock probe단점을 개선시킨 FISSEQ (fluorescent in situ sequencing)방법이 개발되었다. 이에 따라 Seurat, DistMap, novoSpaRc와 같은 computational 기술 역시 함께 발전하고 있으며, spatial에서 세포의 progression 및 기능을 이해하고자 하는 노력이 계속되고 있다[21, 22].
2-5. Single cell multi omics
Single cell sequencing을 이용한 연구는 하나의 세포에서 유전체, 후성유전체, 전사체, 단백체 등의 오믹스를 동시에 분석할 수 있는 방향으로 발전하고 있다. 유전체와 전사체 정보를 통합함으로써 DNA copy number variation이 유전자 발현에 미치는 영향, 유전체 변화에 따른 전사체의 변화, coding 또는 noncoding 부위의 mutation이 transcript 발현 수준에 주는 영향을 확인할 수 있다[23]. G&T-seq(genome & transcriptome sequencing)은 WGA(whole genome amplification)기술과 Smart-seq2를 결합시킨 기술로, 단일 세포로부터 얻은 유전체와 전사체 정보를 통합하여 대규모의 copy number variation(CNV)과 CNV 지역에서의 전사 수준 간의 상관관계를 직접적으로 확인할 수 있게 하였다[24]. 이러한 단일 세포 수준에서의 유전체와 전사체의 통합은 genome variation을 cell heterogeneity의 영향 없이 transcriptional variation에 직접 연결시켜 주며, DNA와 RNA에서 발견되는 돌연변이는 서로 연관되어 있어 동일한 세포의 DNA와 RNA를 봄으로써 더 높은 정확도로 DNA 돌연변이를 확인할 수 있게 한다.
DNA methylation과 전사체의 관계가 매우 중요하게 여겨지면서 이를 단일 세포에서 함께 분석하는 scM&T-seq(single cell methylome and transcriptome sequencing)이 개발되었고, 이는 단일 세포 간의 DNA methylation 차이와 gene transcription variance의 관계 연구를 가능하게 하였다[25]. 이 뿐만 아니라 염색질에 대한 정보와 전사체를 통합시킨 기술도 등장하였다. 단일 세포 전사체 분석 기술인 sci-RNA-seq과 후성유전체 분석 기술인 sci-ATAC-seq을 하나의 protocol로 통합시킨 sci-CAR (single cell combinatorial indexing-chromatin accessibility and mRNA)는 세포 유형마다 다르게 발현하는 유전자와 그것을 조절하는 염색질 부위의 관계를 확인할 수 있게 하였다[26].
RNA와 단백질은 직접적인 생물학적 연관성을 가지고 생물학적 시스템의 특성을 결정하지만, mRNA와 단백질의 서로 다른 반감기, 전사 후 변형에 의한 영향 등으로 인해 mRNA와 단백질 수준 사이의 상관관계 확인에 어려움이 있었다[23]. 단일 세포 수준의 전사체와 단백체의 동시 확인은 RNA와 단백질의 abundance를 탐구할 수 있게 하여 이러한 어려움을 해결하는데 도움을 주고 있다. 단일 세포수준에서 epitope와 전사체를 cellular indexing하는 CITE-seq과 REAP-seq은 oligonucleotide-labeled antibody로 세포 단백질과 전사체를 동시에 분석하는 기술로, 수십 개의 항체로 barcoding된 단백질과 20,000개 이상의 유전자를 한번에 detection할 수 있다[17, 18].
이와 같이 유전체, 후성유전체, 전사체, 단백체를 단일세포에서 통합적으로 분석함으로써, 세포의 heterogeneity 영향을 받지 않으면서 전사체 단일 연구 시 발생하는 한계를 극복하고자 하는 노력이 진행되고 다양한 기술이 개발되었다(Table 1). Single cell multi-omics를 통해 multiple dataset을 통합하여 새로운 biological process를 밝힐 수 있으며, 서로 다른 오믹스 데이터 간의 상관관계 분석 등 단일 세포의 상태를 보다 포괄적으로 묘사할 수 있다. 지속적으로 발전하는 bioinformatics 알고리즘과 계산법, 실험적 기술 발전을 통해 점점 복잡하고 중요한 분석이 가능해질 것이다. 그리고 이러한 single cell multi-omics는 heterogeneous한 세포 집단으로부터 세포의 subtype을 보다 정확히 구분하는데 도움이 될 수 있다[27]. 세포 유형 특이적으로 조절되며 보다 안정한 후성유전적 변화를 scRNA-seq을 통해 얻은 transcript 발현 수준에 대한 정보와 통합하면 scRNA-seq에서 발견하지 못했던 세포의 subtype을 발견할 수 있다. 또한 세포 증식과 분화 시 일어나는 유전자 발현 변화를 확인할 수 있는 전사체와, 세포 분열 시 얻어지고 딸세포로 전달되는 후성유전적 변화에 대한 정보를 통합하면 보다 정확한 lineage trajectory가 가능하다[2]. 현재 single cell multi-omics 데이터로 서로 다른 오믹스 정보 간의 상관관계를 밝힐 수 있으며, 기술이 발달함에 따라 오믹스 간의 인과관계를 밝히는 데에도 효과적일 것으로 기대된다.
Methods |
Measurement |
References |
G&T-seq (genome & transcriptome sequencing) |
genome, mRNA transcriptome |
Macaulay et al., 2015 |
scM&T-seq (single cell methylome&transcriptome sequencing) |
DNA methylome, mRNA transcriptome |
Angermueller et al., 2016 |
sci-CAR (single cell combinatorial indexing-chromatin accessibility and mRNA) |
mRNA transcriptome, chromatin accessibility |
Cao et al., 2018 |
CITE-seq (Cellular Indexing of Transcriptomes and Epitopes by Sequencing) |
mRNA transcriptome, proteome |
Stoeckius et al., 2017 |
REAP-seq (RNA expression and protein sequencing assay) |
mRNA transcriptome, proteome |
Peterson, V.M., et al., 2017 |
ScTrio-seq (single cell triple omics sequencing) |
genome, DNA methylome, mRNA transcriptome |
Hou et al., 2016 |
scNMT-seq (single-cell nucleosome, methylation and transcription sequencing) |
DNA methylation, Nucleosome, mRNA transcriptome |
Clark et al., 2018 |
NOMe-seq (nucleosome occupancy and methylome sequencing) |
nucleosome, DNA methylome |
Kelly et al., 2012 |
Table 1. Single cell multi-omics methods
결론
scRNA-seq은 세포의 heterogeneity에 영향을 받지 않고 세포 유형을 구분하여 분석할 수 있을 뿐만 아니라 동일한 cell에서도 다르게 발현하는 유전자 양상을 볼 수 있으며, progenitor단계에서 분화되는 과정에서의 유전자 변화를 확인하여 lineage tracing을 가능하게 한다. 그러나 아직까지 여러 가지 해결해야 할 문제점도 남아 있다. scRNA-seq은 적은 양의 transcript를 증폭시켜 분석하기 때문에 noise가 많고, 높은 dropout event로 인해 missing gene measurement가 발생하여 데이터 분석에 어려움이 있다. 증폭 과정에서 생기는 noise는 UMI(unique molecular barcode)로 해결할 수 있으며, RNA capture efficiency를 높여 noise를 감소시키려는 노력도 진행중이다. 단일 세포 분리과정이나 sequencing library를 만드는 과정에서 서로 다른 protocol을 사용하여 발생하는 batch effect도 분석에 어려움을 주는데, 이를 해결하기 위해 batch effect를 감소시키는 computation 방법들이 개발되었으며 서로 다른 샘플을 함께 multiplexing하는 방법으로도 이를 감소시킬 수 있다. 이 외에도 조직의 특성, 세포 별 크기 및 digestion condition에 대한 반응이 달라 발생하는 cell composition bias도 해결해야 하는 요인으로 남아있다.
전사체 뿐만 아니라 유전체, 후성유전체, 단백체 등 다양한 분자 유형을 세포 유형, 상태를 구분하여 분석할 수 있는 다양한 single cell sequencing기술이 개발되었고, 단일 세포 수준의 오믹스 데이터를 통합하여 분석하는 multi-omics의 발전은 유전자와 복합적으로 얽혀 질병을 발생시키는 요인들과 그들의 상호작용을 분석할 수 있게 하여 단일 오믹스 데이터에서 오는 한계를 극복하는데 도움을 주고 있다 (Figure 1). 이러한 단일 세포 다중 오믹스 분석은 scRNA-seq을 도와서 세포 다양성을 기술하고, lineage tracing의 정확도를 높이며, 새로운 세포 유형을 확인할 수 있게 해준다. 또한 다양한 오믹스 정보를 동일한 세포에서 관찰함으로써 오믹스 간의 조절 메커니즘을 보다 직접적으로 볼 수 있는 기회를 제공한다[2, 6]. 향후 single cell 분석의 문제점이 점차 해결되고 분석 기술의 발달이 이루어지면 질병 연구, lineage tracing 등 다양한 분야에서 단일 세포 수준의 RNA sequencing의 비중이 높아지고 많은 연구에서 bulk RNA를 대체할 것으로 예상된다. 궁극적으로는 scRNA-seq을 통해 환자 개개인의 세포 종류의 구성, 유전자 발현을 분석함으로써 맞춤 의학의 발전에 기여할 수 있을 것이다.
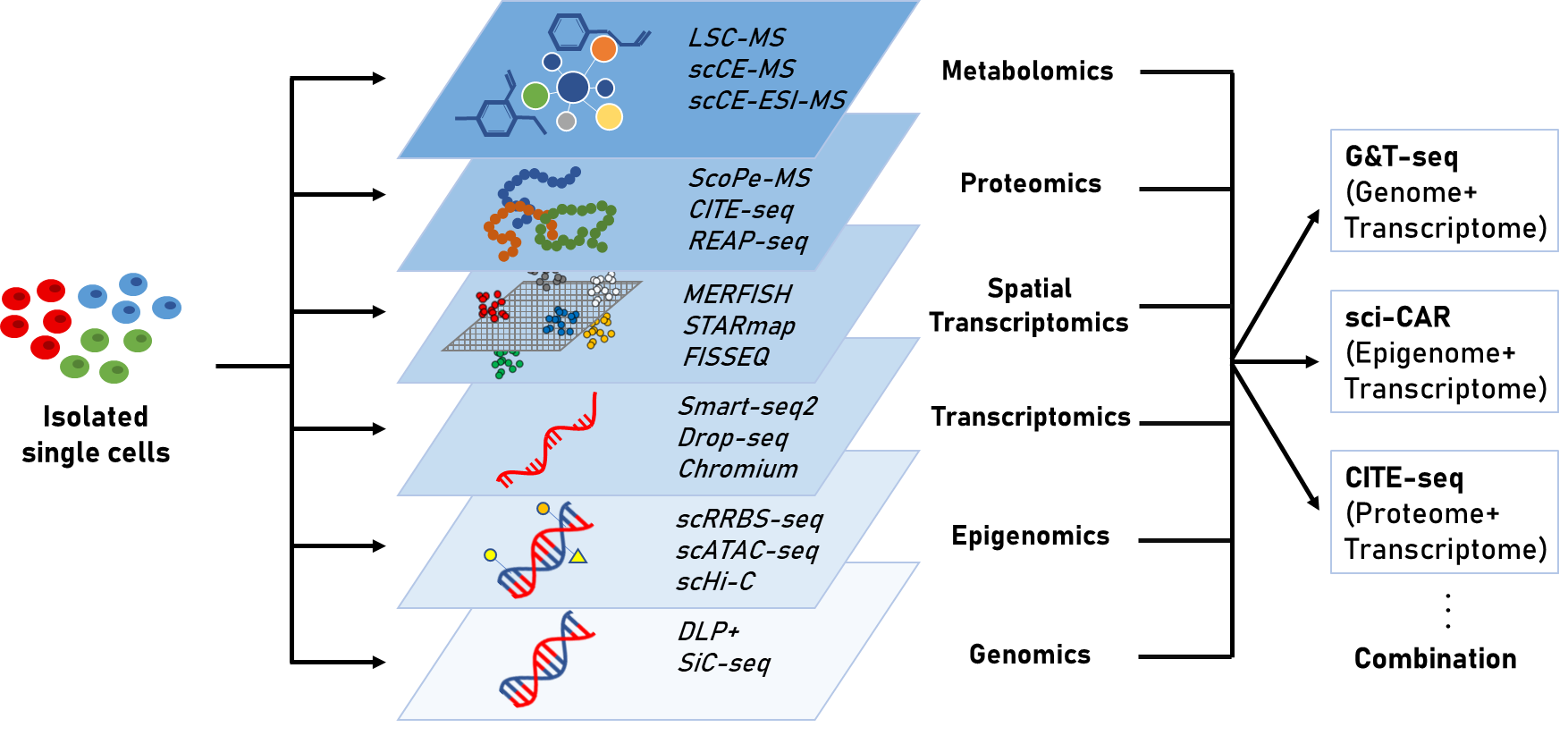
Figure 1. multiple molecular type에서의 single cell sequencing과 multi-omics
참고 문헌
1. Pennisi, E., Development cell by cell. 2018, American Association for the Advancement of Science.
2. Hu, Y., et al., Single cell multi-omics technology: methodology and application. Frontiers in cell and developmental biology, 2018. 6: p. 28.
3. Yeung, E.S., Genome‐wide correlation between mrna and protein in a single cell. Angewandte Chemie International Edition, 2011. 50(3): p. 583-585.
4. Wang, Y. and N.E. Navin, Advances and applications of single-cell sequencing technologies. Molecular cell, 2015. 58(4): p. 598-609.
5. Ramsköld, D., et al., Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature biotechnology, 2012. 30(8): p. 777.
6. Song, Y., et al., Single cell transcriptomics: moving towards multi-omics. Analyst, 2019. 144(10): p. 3172-3189.
7. Han, X., et al., Mapping the mouse cell atlas by microwell-seq. Cell, 2018. 172(5): p. 1091-1107. e17.
8. Cao, J., et al., Comprehensive single-cell transcriptional profiling of a multicellular organism. Science, 2017. 357(6352): p. 661-667.
9. Wu, H., et al., Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. Journal of the American Society of Nephrology, 2019. 30(1): p. 23-32.
10. Guo, H., et al., Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nature protocols, 2015. 10(5): p. 645.
11. Rotem, A., et al., Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nature biotechnology, 2015. 33(11): p. 1165.
12. Buenrostro, J.D., et al., Single-cell chromatin accessibility reveals principles of regulatory variation. Nature, 2015. 523(7561): p. 486.
13. Nagano, T., et al., Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature, 2013. 502(7469): p. 59.
14. Orecchioni, M., et al., Single-cell mass cytometry and transcriptome profiling reveal the impact of graphene on human immune cells. Nature communications, 2017. 8(1): p. 1109.
15. Goltsev, Y., et al., Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell, 2018. 174(4): p. 968-981. e15.
16. Budnik, B., et al., SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome biology, 2018. 19(1): p. 161.
17. Stoeckius, M., et al., Simultaneous epitope and transcriptome measurement in single cells. Nature methods, 2017. 14(9): p. 865.
18. Peterson, V.M., et al., Multiplexed quantification of proteins and transcripts in single cells. Nature biotechnology, 2017. 35(10): p. 936.
19. Wang, G., J.R. Moffitt, and X. Zhuang, Multiplexed imaging of high-density libraries of RNAs with MERFISH and expansion microscopy. Scientific reports, 2018. 8(1): p. 4847.
20. Eng, C.-H.L., et al., Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature, 2019. 568(7751): p. 235.
21. Wang, X., et al., Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science, 2018. 361(6400): p. eaat5691.
22. Lee, J.H., et al., Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nature protocols, 2015. 10(3): p. 442.
23. Macaulay, I.C., C.P. Ponting, and T. Voet, Single-cell multiomics: multiple measurements from single cells. Trends in Genetics, 2017. 33(2): p. 155-168.
24. Macaulay, I.C., et al., G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nature methods, 2015. 12(6): p. 519.
25. Angermueller, C., et al., Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nature methods, 2016. 13(3): p. 229.
26. Cao, J., et al., Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science, 2018. 361(6409): p. 1380-1385.
27. Luo, C., et al., Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science, 2017. 357(6351): p. 600-604.
첨부파일